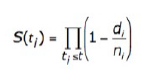GDC FAQs
-
From what data are mutation-based visualization features derived?
The mutation-based visualization features are derived from open-access MAF files produced by GDC variant-calling pipelines.
-
Why are there some projects without data analysis and visualization features?
Data analysis and visualization features are only available for projects which maintain open-access MAF files. Programs such as the Molecular Analysis for Therapy Choice (MATCH) trial maintain controlled-access MAF files only. As such, data analysis and visualization cannot be applied to MATCH projects.
-
In the Most Frequent Mutations table for the VEP impact score, which algorithm in the VEP is the GDC using to determine “H" or “M”?
The IMPACT is categorized by the Sequencing Ontology type of the variants that is also compatible to snpEff. The VEP IMPACT rating is a separate rating given for compatibility with other variant annotation tools (e.g. snpEff). Basically, each category is associated with a set of SO terms:
- HIGH: The variant is assumed to have high (disruptive) impact in the protein, probably causing protein truncation, loss of function or triggering nonsense mediated decay: transcript_ablation, splice_acceptor_variant, splice_donor_variant, stop_gained, frameshift_variant, stop_lost, start_lost, transcript_amplification
- MODERATE: A non-disruptive variant that might change protein effectiveness: inframe_insertion, inframe_deletion, missense_variant, protein_altering_variant, regulatory_region_ablation
- LOW: Assumed to be mostly harmless or unlikely to change protein behavior: splice_region_variant, incomplete_terminal_codon_variant, stop_retained_variant, synonymous_variant
- MODIFIER: Usually non-coding variants or variants affecting non-coding genes, where predictions are difficult or there is no evidence of impact: coding_sequence_variant, mature_miRNA_variant, 5_prime_UTR_variant, 3_prime_UTR_variant, non_coding_transcript_exon_variant, intron_variant, NMD_transcript_variant, non_coding_transcript_variant, upstream_gene_variant, downstream_gene_variant, TFBS_ablation, TFBS_amplification, TF_binding_site_variant, regulatory_region_amplification, feature_elongation, regulatory_region_variant, feature_truncation, intergenic_variant
Details about predicted data in variations are available at ENSEMBL
-
How is survival analysis calculated?
Survival analysis in the GDC uses a Kaplan‑Meier estimator:
- S(t ) is the estimated survival probability for any particular one of the t time periods
- n is the number of subjects at risk at the beginning of time period t
- d is the number of subjects who die during time period t
Please refer to the GDC Data Portal User's Guide Projects for additional information. -
Can I use the GDC Application Programming Interface (API) to retrieve data sets associated with visualizations?
Yes. The GDC provides additional analysis endpoints to retrieve data sets associated with visualizations. Analysis endpoints include: survival, top_cases_counts_by_genes, top_mutated_genes_by_project, top_mutated_cases_by_gene, top_mutated_cases_by_ssm, and mutated_cases_count_by_project.
Please refer to the GDC API User's Guide Analysis Section for additional information.
-
Where can I find information on the format of GDC MAF Files?
Please refer to the GDC MAF File Specification to obtain detailed information on the format of GDC MAF files.
-
On the GDC Project summary page or Mutation Frequency > Gene tab, why are the # of Mutations sometimes less than the # Affected Cases?
The “# Mutations” column in the Project or Mutation Frequency > Gene tab displays the number of distinct (unique) mutations within the affected cases and not necessarily the total number of all mutations within the project or query filter.
-
Why does the GDC display common genes such as TTN that are associated with every cancer in the most frequently mutated genes table?
The GDC is not normalizing frequency by gene length. This is currently under discussion. As such, these genes are appearing in the mutated genes table. Users can filter by the COSMIC Cancer Gene Census to display only genes for which mutations have been causally implicated in cancer.
-
How do I search for a particular mutation?
To search for a mutation, you can utilize the Quick Search bar at the top right portion of the GDC Portal by entering in either a dbSNP reference cluster ID (rs#) or the coordinates of the chromosomal change. For example entering in 'rs121912651' or 'chr17:g.7674221G>A' will bring the user to the mutation entity page for that mutation.
-
What considerations should be taken when comparing samples with different target capture kits?
Target capture kits are used to "target" specific regions of a given genome for the Whole Exome Sequencing (WXS) and Targeted Sequencing experimental strategies. Users should therefore take care when comparing data from different target capture kits for the WXS and Targeted Sequencing experimental strategies because of potential differences in genomic regions targeted, variant filtering, and subsequent variants recovered. Additionally, users should also consider that Whole Genome Sequencing (WGS) does not use target capture kits and may thus recover variants that are excluded in the WXS and Targeted Sequencing experimental strategies.
From the GDC Data Portal "Repository" tool, the names of the target capture kits can be accessed by clicking the "Add a Custom Filter" link and choosing "analysis.metadata.read_groups.target_capture_kit" from the menu. The GDC does not distribute target or bait files because they are intellectual property. Users should contact each individual project directly if detailed target capture region information is needed.
-
Does the GDC correct for batch effects across samples?
The GDC does not perform batch effect corrections across samples for the following reasons:
- The GDC accepts new data on an ongoing basis and has continuous data processing and frequent release plans. It is operationally difficult to perform cross-GDC batch effect corrections in every release.
- Many of the batch effect correction processes require manual considerations and project-specific knowledge.
- Automatic batch effect correction might remove real biological meaningful signals, especially when batch effect is confounded with real biological effects.
As such, the GDC prefers that users to perform their own batch effect removal.
-
Does the GDC use common exomes across all whole exome platforms?
There is a variety of target capture kits used by different sequencing centers. Most of the whole exome capture kits share many common genomic regions, especially for cancer related genes; However, which exons are included is totally dependent on the vendor's library preparation kit. There are often more differences among capture regions from different Targeted-Sequencing/Panel data.
The name of the capture kit used is available from the GDC read group properties. However, the GDC does not distribute the BED files for the read groups associated with these capture kits because some of them are proprietary.
-
How often does the GDC update the workflow/reference genome? If the GDC updates the workflow/reference genome, does the GDC re-process all data sets?
For the reference genome, the GDC has been using an augmented version of GRCh38.p2 (with additional decoy sequences and virus sequences) since inception. The GDC does not use alternative contigs, and only derives high-level data from the major chromosomes, so the same reference genome is used for both gene model GENCODE v22 (from Data Release 1 to 31) and GENCODE v36 (from Data Release 32). As future versions of the reference genome are released, e.g., GRCh39, the GDC will evaluate the benefits of updating data to utilize the new version. By updating the reference genome, the GDC would expect to re-process all data sets. For information on the reference genome used by the GDC, please refer to the GDC Reference Files.
For workflow updates, the GDC prefers to keep the workflow stable, and will not update unless there are necessary updates such as updates of the reference genome or gene model, or major algorithm updates in the tools that could result significant changes in the generated data. When workflow updates are actually needed, the GDC categorizes them as either major updates or minor updates depending on whether the update significantly affects the output data. The GDC will re-process all existing data sets in major workflow updates, and such examples include transitioning the RNA-Seq genomic BAM alignment workflow into a new version that generates three BAMs and STAR counts; and updating the MAF workflow to add additional functions to the MAF files. Minor updates mostly happen to resolve bugs, security issues, and/or compatibility issues. For example, the GDC DNA-Seq alignment workflow has been updated several times to address quality issues from various submitted data; however, because the main alignment algorithm remains almost the same, the GDC does not need to re-process all the data sets for these minor updates.
-
Why did the GDC remove HTSeq for gene expression quantification?
HTSeq had been the default RNA-Seq expression quantification tool since the first GDC data release. The GDC later updated the RNA-Seq alignment and quantification workflow to include STAR Count, which generates stranded counts by default in addition to the existing unstranded counts. During Data Release 32 for gene model updates, the GDC had 1) augmented the existing STAR Count output to include FPKM and FPKM-UQ normalizations; 2) reprocessed all the TCGA data using the latest RNA-Seq workflow with STAR Count. Because both tools use very similar counting strategies, and STAR Count has the advantages in both running time and the additional stranded counts, the GDC removed HTSeq workflow in Data Release 32.
-
Does the GDC provide access to germline variants?
Any germline SNP calls are not available for exploration in the GDC Data Portal. Instead, alignments for germline data are available under controlled access. Users with appropriate access may use the alignments to generate germline variants.
Some somatic variants callers, such as MuTect2, also output somatic calls with some level of germline possibilities, such as those labelled as "germline_risk". Please note that these calls are, by no means, germline variants. They are somatic calls with boundary probability of germline risks.
-
Why did the GDC remove SomaticSniper?
The SomaticSniper whole exome variant caller was one of the first generation somatic mutation callers developed by the scientific community. It works the best with blood cancer that has high level of tumor-in-normal contaminations, but is often overly permissive for solid tumors. Since our first data release in 2016, the GDC has gradually adopted newer tools or new tool versions, and has transited the focus of somatic variant calling from any single caller to multi-caller ensemble.
After comparing ensemble calls with and without SomaticSniper and also receiving feedback from the authors of SomaticSniper, the GDC decided to remove this tool from our production in Data Release 35. The GDC still maintains other four whole exome variant callers, including MuSE, MuTect2, Pindel, and VarScan2.
-
Why do some projects with WGS structural variant data have BEDPE files and some projects do not?
Generally any WGS data should have associated structural variant files (BEDPE) except in the cases in which either there are no tumor/normal matches or when variant calling has not been implemented yet.
-
In the GDC Data Portal, where is the histogram of top frequently mutated genes for a cohort?
To view the top frequently mutated genes for a cohort, first build a cohort using the Cohort Builder and then select the Mutation Frequency tool in the Analysis Center
-
Why do some genes show no expression in STAR results across all samples, even though I can see mapped reads in the raw RNA-Seq data?
STAR gene expression quantification excludes reads that are mapped to multiple different genes. This can cause some genes to appear with zero expression in the final counts, even if mapped reads are present in the raw data.
One common reason for this is gene overlap. These genes often have their exons entirely encompassed within other genes, and in such cases, STAR cannot assign reads to them because they are ambiguous. To check if a gene falls into this category, you can refer to the following lists: Stranded Counting Overlap Gene List: overlap.gene.stranded.tsv, and Strandless Counting Overlap Gene List: overlap.gene.strandless.tsv.
-
How does the GDC choose the default transcript for each variant?
When a mutation overlaps multiple transcripts or genes, the GDC annotates all consequences in the all_effects column of the MAF file and in the CONSEQUENCE table on the Mutation Summary Page. One transcript is then selected as the default for detailed annotation and visualization where a single consequence is shown.
The default is chosen based on annotations from the Variant Effect Predictor (VEP), prioritizing the most severe consequence on the most impactful transcript biotype (See: selected annotation for the 'OneEffect'). The GDC also applies a curated transcript override file from MSKCC, which defines preferred transcripts for key genes, along with consideration for canonical and longest transcripts.
During GENCODE v36 updates in GDC Data Release 32 (DR 32), Some hotspot mutations may display a different default consequence annotation. For example, BRAF V600E is shown as BRAF V640E after DR32 because the curated BRAF transcript ENST00000288602 was updated by GENCODE with an addition of 40 amino acids at the N-terminus. Changes like this affect the default consequence annotations for all BRAF mutations and may similarly impact other genes with updated transcript models. Although the default annotation changed, users can still find V600E listed in the all_effects column in MAF files or the CONSEQUENCE table on the Mutation Summary Page alongside other transcript annotations.
-
How are the five categories of copy number changes determined?
The GDC begins with integer-level estimates of absolute copy number generated by either the ASCAT or ABSOLUTE pipeline. To establish a baseline, an integer-valued sample ploidy is computed as follows:
- For gene-level CNV, the mode of copy number values is used across all autosomal protein-coding genes.
- For segment-level CNV, a length-weighted mode of copy number values is computed across all autosomal segments.
- In cases of a tie, the mode is rounded up.
- Please note that the integer-valued sample ploidy used here differs from the floating-point ploidy estimates produced directly by the ASCAT or ABSOLUTE pipelines. The latter should be considered the more precise representation and is recommended for use in most other bioinformatics analyses.
Based on this sample ploidy value, the GDC assigns copy number categories as:- Homozygous deletion: copy number = 0 Loss: 0 < copy number < sample ploidy
- Neutral: copy number = sample ploidy
- Gain: sample ploidy < copy number < 2 × sample ploidy
- Amplification: copy number ≥ 2 × sample ploidy
-
Why do CNVs of different genes in the GDC Data Portal differ from other genomic portals?
This discrepancy is due to differences in the data processing pipelines used by the GDC and other genomic portals. At the GDC, gene-level CNVs are derived from a mix of standardized pipelines. For TCGA projects, the CNV values are prioritized in the following order: SNP6 ABSOLUTE (LiftOver) > SNP6 ASCAT3 > WGS AscatNGS > SNP6 ASCAT2. All of these workflows produce absolute integer copy number values.
In contrast, data from other genomic portals may come from more diverse sources. The same TCGA project may appear under different studies, with gene-level CNVs derived from the original publication segment mean values or data ingested directly from the GDC. The exact data origin and processing steps in other genomic portals can vary by study.