The data model is designed to maintain data and metadata consistency, integrity, and availability while accommodating:
- Biospecimen, clinical, and cancer genomic data and metadata
- Multiple, disparate NCI ongoing projects
- Completely new, as yet unthought of projects
- Ongoing changes and technological progress
- Frequent and complex queries from both external users and internal administrators
To meet these requirements, the design and implementation of the data model leverages:
- Flexible but robust graph-oriented data stores
- Indexed document stores for API and front end performance
- Ontology-based concept and data element definition
- Schema-based entity and relationship validation on loading
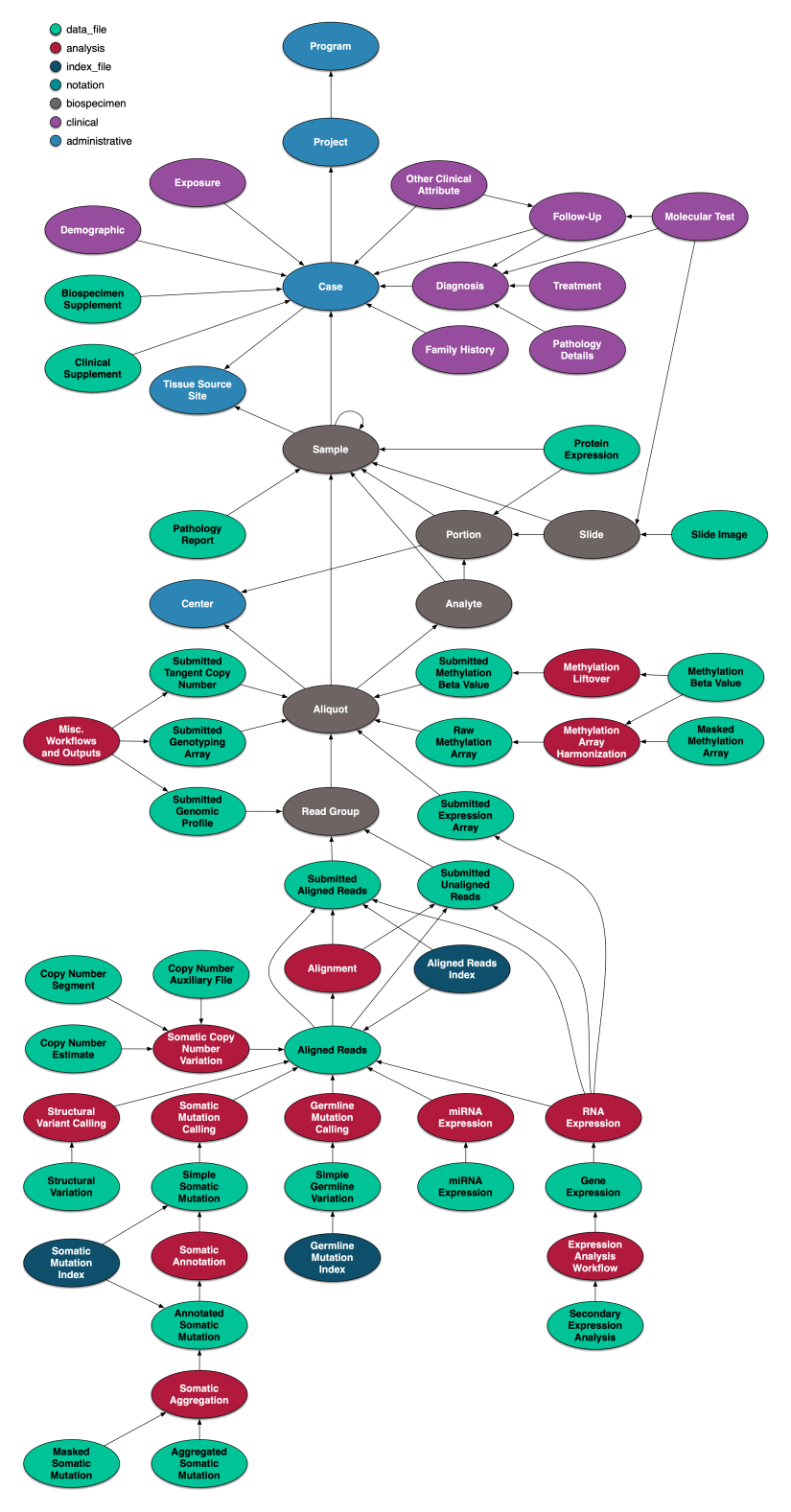
Graph Representation of the GDC Data Model
The GDC data model is represented as a graph with nodes and edges, and this graph is the store of record for the GDC. It maintains the critical relationship between projects, cases, clinical data and molecular data and insures that this data is linked correctly to the actual data file objects themselves, by means of unique identifiers. The graph is designed in terms of the "property graph" model, in which nodes represent entities, edges between nodes represent relationships between entities, and properties on both nodes and edges represent additional data which describe entities and their relationships. Relationships are encoded as edges of a given type which associate exactly two nodes. Properties of nodes or relationships are sets of key-value pairs.
Original metadata as submitted by external users is extracted and loaded first into the graph. Representations of the data provided by the other GDC components are derived from the authoritative graph model. Note that file and archive objects are not stored in the graph, but rather in an external object store. The node/edge structure of the graph is depicted below.
Data Dictionary and Validation
Item semantics (names, accepted values) and their interrelationships can be changed or updated easily within the GDC data model. However, they cannot be completely free to vary at any time, since users depend on the stability of the graph and its vocabulary. Therefore, a means to encode a schema for the metadata is also an integral part of the GDC data model. The GDC maintains standard terms, their definitions and references to public ontologies and vocabularies in dictionary files. Dictionaries are expressed in YAML, utilizing JSON Schema conventions to be computable and are intended to provide both the internal GDC standard vocabulary and the publicly-accessible source of GDC term information. Each node is represented by a dictionary file that specifies node properties and values, as well as allowable edges to other node types. Dictionaries are kept in version control with periodic releases.
As submitted metadata is processed, the associations among cases, biospecimens, and data files are extracted and stored in the graph. To prevent errors in those associations from entering the graph, a validation system is implemented at the application level. Primary validation rules (such as data type and accepted value checking) are defined in JSON Schema as part of the GDC data dictionaries. Secondary validations (those that, for example, confirm data consistency among values of different entries in a submission) are implemented as custom modules and referenced in relevant dictionaries. Validation is executed on incoming metadata and must pass before they are allowed to enter the graph itself.
Data Type and Subtype Definition and Tagging
To categorize files from legacy programs on initial import, GDC developed and implemented a system of associating data type, data subtype, platform, experimental strategy, and access type assignments to files based on file name pattern matching. To facilitate user search and download of desired data, a system of short tokens or tags has also been developed to classify externally available files into related groups outside the set of defined data typing dimensions listed above. The data typing facets and tags provide the basis for categorizing files submitted or generated in the future. See the TCGA Tags Guide for more details.