A summary of the many tools, applications, and other resources the GDC provides for retrieving, downloading, and analyzing data in the GDC, submitting data to the GDC, and processing data through GDC bioinformatics pipelines.
| Image | Resource |
|---|---|
|
|
GDC Data PortalThe GDC Data Portal is a robust web-based platform that allows users to search, analyze, and download data from cancer genomic studies. The GDC Data Portal centers around the idea of building cohorts, or groups of cases, before analyzing or downloading data. Users use the Cohort Builder to filter on the rich set of GDC clinical, biospecimen, and available data elements to create custom cohorts for analysis in the Analysis Center. The Analysis Center provides interactive analysis tools supporting gene variant-level analyses and clinical examination. Data is available for download in the Repository. Launch the GDC Data Portal | User's Guide | GitHub Repository |
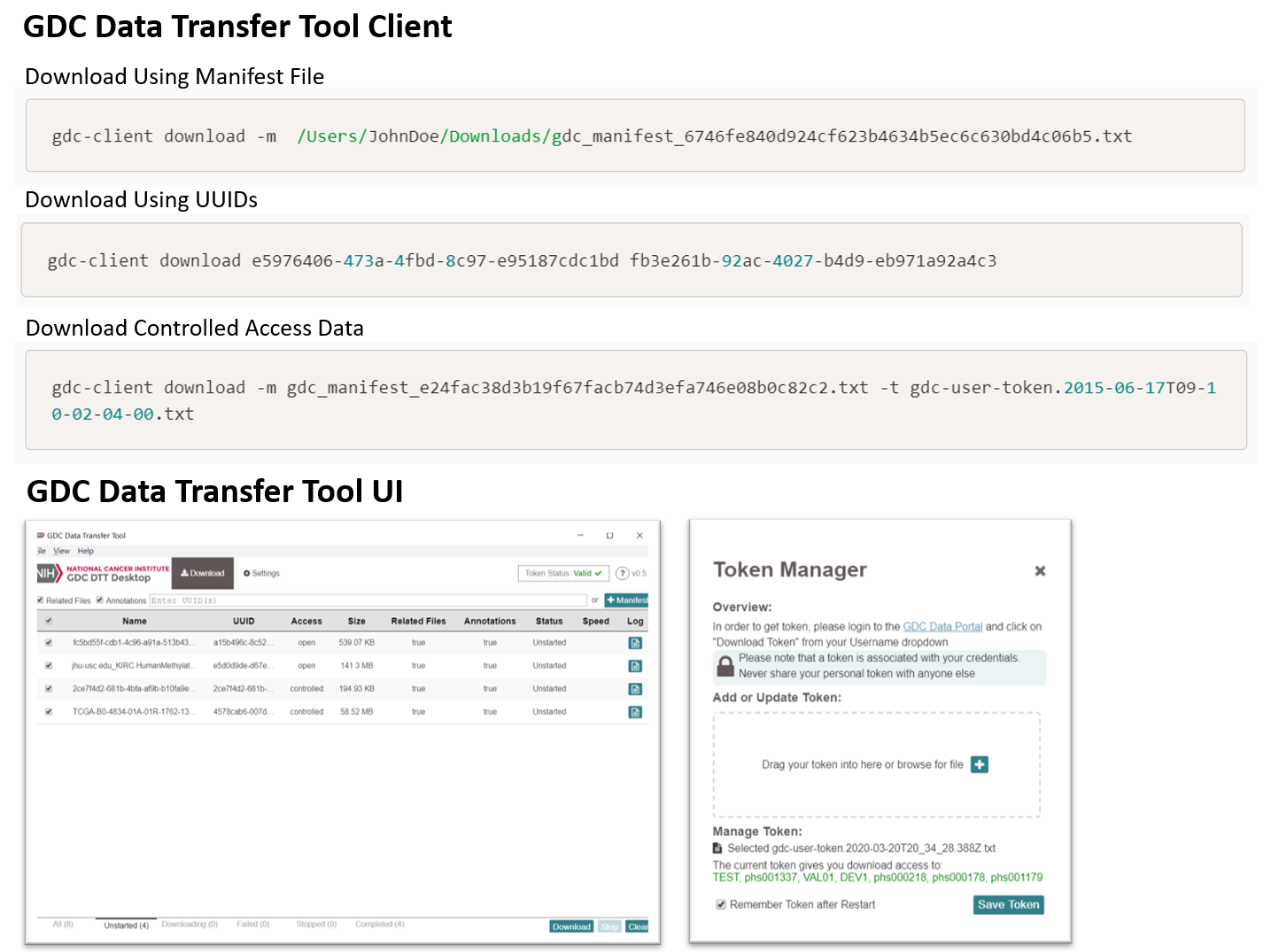
GDC Data Transfer Tool (DTT)The GDC DTT is a command-line driven application for the download and upload of large, high volume data. The GDC DTT provides an optimized method for transferring data to-and-from the GDC and enables resumption of interrupted transfers. The GDC DTT Client provides a command-line interface supporting both GDC data downloads and submissions. The GDC DTT User Interface (UI) provides a user-friendly interface to the GDC DTT Client for downloading data from the GDC. Download the GDC DTT Client and UI | User's Guide | GitHub Repository (Client) |
|
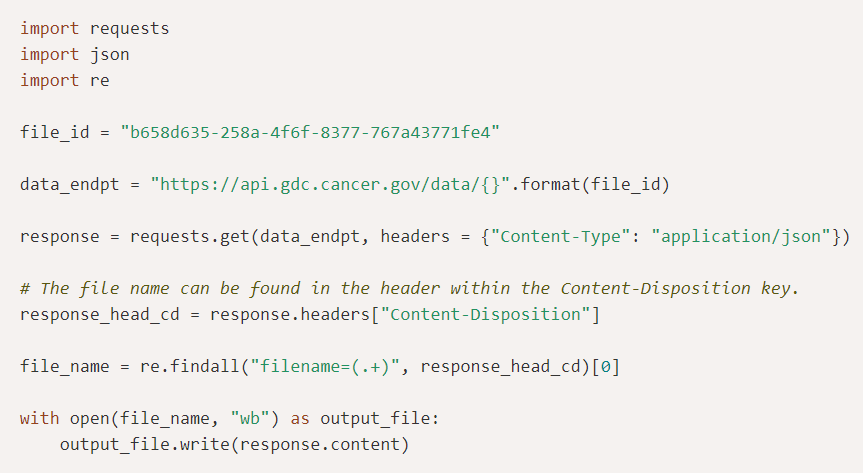
GDC Application Programming Interface (API)The GDC API is a programmatic interface for searching, downloading, submitting, and analyzing GDC data and metadata. The GDC API is the external facing REpresentational State Transfer (REST) interface for the GDC and uses JSON as its communication format, and standard HTTP methods (GET, PUT, POST and DELETE). |
|
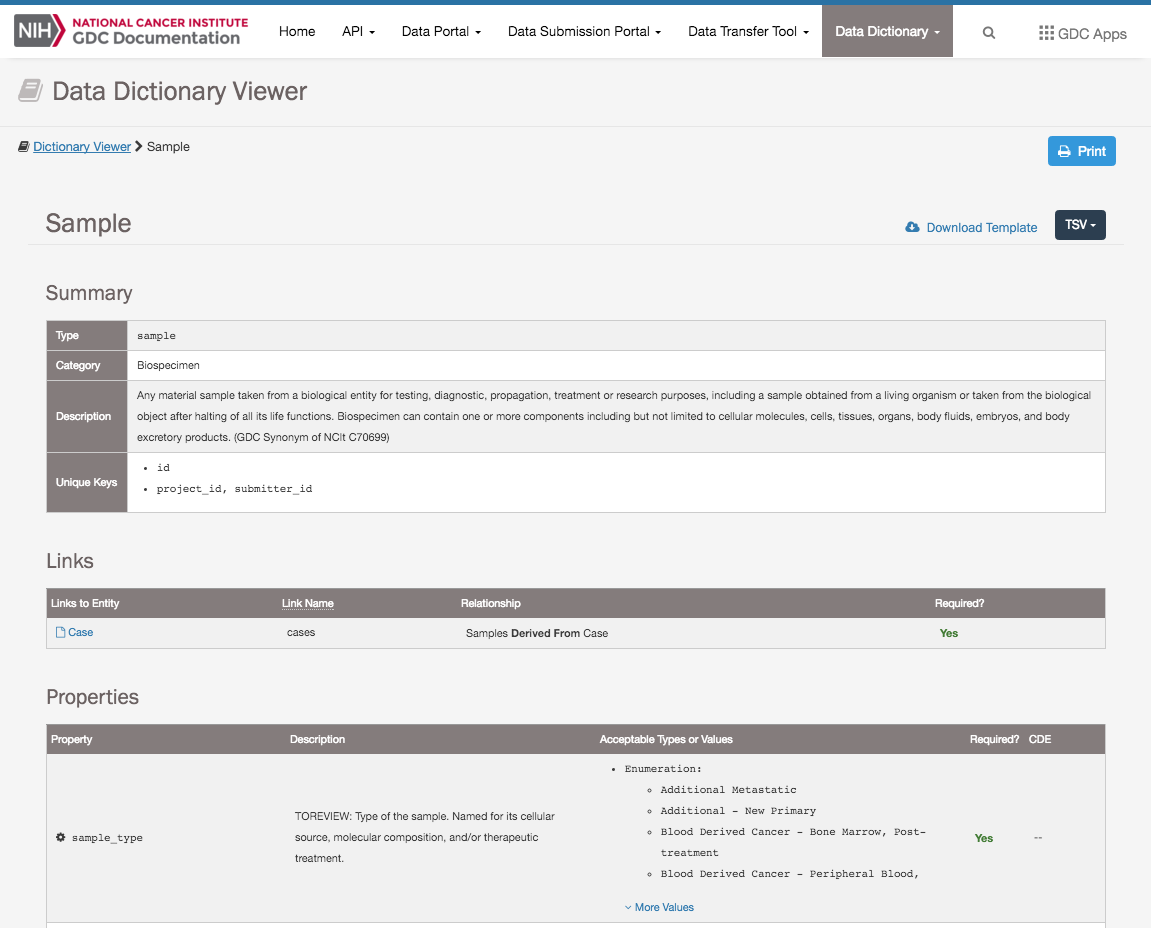
GDC Data Dictionary and Data ModelThe GDC Data Dictionary is a resource that describes the clinical, biospecimen, administrative, and genomic metadata that can be used in parallel with the genomic data generated by the GDC. The dictionary defines the structure of the GDC graph-based data model and the rules the data need to follow. In addition, the dictionary includes information about the relationships between entities within the data model. View the GDC Data Dictionary | GDC Data Model | GitHub Repository (Dictionary) |
|
|
|
GDC Data Submission PortalThe GDC Data Submission Portal is a web-based tool for submitting clinical, biospecimen, and molecular data associated with projects that are registered in dbGaP and accepted for submission into the GDC. Submitted data is validated using built-in GDC review/QC tools. Request Data Submission | GDC Data Submission Processes and Tools | Launch the GDC Data Submission Portal (Login Required) | User's Guide |
|
|
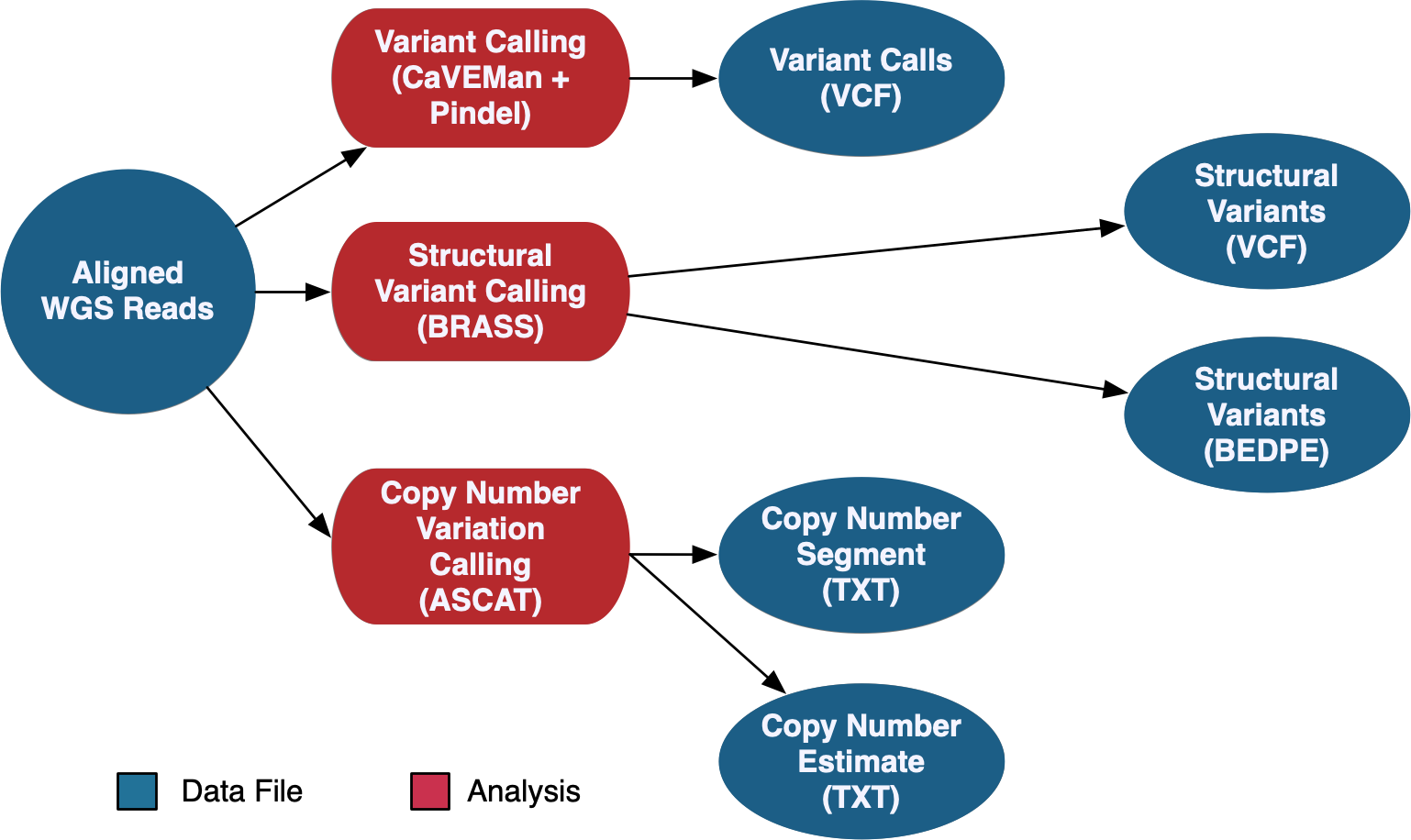
GDC Bioinformatics PipelinesGDC Bioinformatics Pipelines are standard workflows supporting DNA, RNA, and miRNA alignments against a common reference genome (GRCh38) and higher level data generation of these and other data types. View GDC Bioinformatics Pipelines (DNA-Seq, RNA-Seq, miRNA-Seq) | GitHub Repository (GDC Workflows) | Reference Files Used by GDC Pipelines |
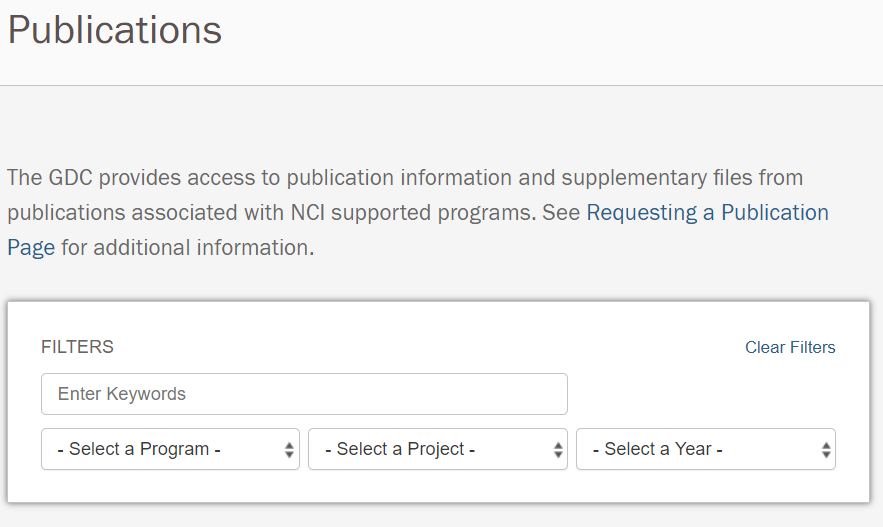
GDC Publication PagesGDC Publication Pages provide access to information and supplementary files from publications associated with NCI supported programs. Search facilities are provided to filter publications by program, project, publication year, and keywords. |